SepsiTest™-UMD: Unbiased rDNA PCR Pathogen Diagnosis |
| 05 November 2018 |
|
Molzym has developed the SepsiTest™-UMD CE IVD system that uses broad-range 16S and 18S rRNA gene PCR and sequencing analysis of eubacteria and fungi, respectively. In this context the removal of host DNA and thereby enrichment of microbial DNA plays an important role for the enhancement of analytical sensitivity (see blog post “MolYsis™ Host DNA Removal – Effect on Pathogen Analysis”). After DNA extraction and PCR (4 hours) a first result on the presence or absence of eubacteria and fungi is obtained. Validation of the results is achieved by supplied extraction and PCR run controls (positive, negative). Amplicons from positive assays are Sanger sequenced, and a BLAST analysis by Molzym’s online tool, SepsiTest™-BLAST, identifies the species on the grounds of a quality-edited library of >7,000 entries of 16S and 18S rRNA genes (see Fig. 1). |
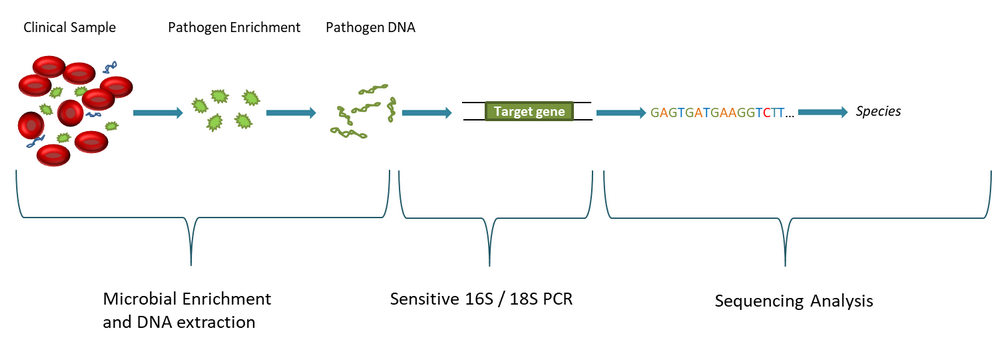 |
| Fig.1: Overview of the unbiased diagnostic approach of SepsiTest™-UMD. |
|
The broad-range identification character of SepsiTest™-UMD has been demonstrated in evaluations including clinical fluid and tissue specimens as well as other materials. The identification of so far 1,359 microorganisms from 303 bacterial and 40 fungal genera emphasizes the unbiased concept of SepsiTest™-UMD diagnosis (inquire the Diese E-Mail-Adresse ist vor Spambots geschützt! Zur Anzeige muss JavaScript eingeschaltet sein!). For further information on clincial evaluations of SepsiTest™-UMD follow the blog “Molecular Diagnostics”. For more detailed information about SepsiTest™-UMD CE IVD please refer to our application note or visit the product site. |
| Application Note Brochure Product Information |

 Different molecular methods have been developed in the last years on the grounds of Real-Time PCR, magnetic resonance, electrospray ionization flight of mass spectrometry, DNA arrays and others for the rapid, culture-independent detection of pathogens. Most available methods are limited to pathogens of specific diseases like sepsis, including S. aureus, E. coli, K. pneumoniae, P. aeruginosa and others. An unbiased approach of diagnosis can detect any pathogen present in a specimen, including besides common pathogens also microorganisms that fail to grow in culture media because of fastidious growth requirements or inhibition as a course of antibiotic.
Different molecular methods have been developed in the last years on the grounds of Real-Time PCR, magnetic resonance, electrospray ionization flight of mass spectrometry, DNA arrays and others for the rapid, culture-independent detection of pathogens. Most available methods are limited to pathogens of specific diseases like sepsis, including S. aureus, E. coli, K. pneumoniae, P. aeruginosa and others. An unbiased approach of diagnosis can detect any pathogen present in a specimen, including besides common pathogens also microorganisms that fail to grow in culture media because of fastidious growth requirements or inhibition as a course of antibiotic.