SelectNA™plus - Robotic Pathogen DNA Extraction System |
| 27 June 2018 |
|
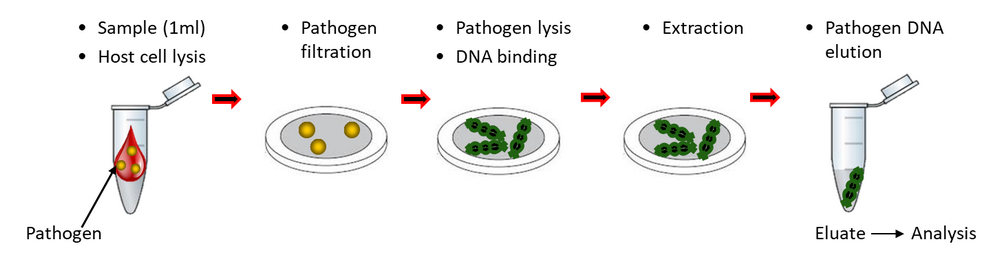
To overcome this problem, Molzym developed a process, MolYsis™, by which host cells are lysed while microbes stay intact and the released human DNA is degraded. In addition, cell-free DNA from microbes that have been digested by the host immune system is degraded. After free DNA degradation, microbial cells are steered into DNA extraction and isolation procedures leading to enriched microbial DNA in the eluate. In other words, only microbial DNA originating from growing and non-growing intact cells (antibiotic-inhibited or fastidious pathogens) can be detected. Positive effects of MolYsis™ treatment on the detection sensitivity of pathogens by qPCR amplification and community analysis methods have been reported [3-6], including gene clone library preparations and Next Generation Sequencing (NGS). |
|
|
|
Molzym offers the MolYsis™ technology fully automated on the SelectNA™plus instrument (Fig. 1). By this, microbial DNA can be enriched and isolated from a great variety of specimens, including blood, urine, aspirates from joints, ascites, cerebrospinal fluid and other sites, wound swabs, tissue biopsies and other clinical materials. The eluted DNA can be used for qualitative and quantitative Real-Time PCR, cloning as well as microbiome analysis by NGS. |
|
SelectNA™plus is a bench-top robotic system (Fig. 2) that processes samples of up to 1 ml volume, including all steps from host cell lysis and human DNA depletion to microbial DNA purification (Fig. 1). For this, the instrument has only to be loaded with cartridges, enzyme vials and elution tubes of SelectNA™plus kits along with the samples. Up to 12 samples can be extracted at a time. Two kits are available for the use with the SelectNA™plus robot: • MolYsis-SelectNA™plus: Microbial DNA extraction for research use – typical applications are fields where host DNA negatively influences assay performances, including development of sensitive PCR and qPCR detection assays of bacterial or fungal loads or microbiome analysis by NGS. • Micro-Dx™ CE IVD: Microbial DNA extraction and broad-range PCR – CE-marked kit for the routine in vitro sequencing diagnosis of bacteria and fungi in clinical samples. For more detailed information of SelectNA™plus applications please refer to our application notes. |
| References |
|
[1] Handschur M, Karlic H, Hertl C, Pfeilstöcker M, Haslberger AG (2009) Preanalytic removal of human DNA eliminates false signals in general 16S rDNA PCR monitoring of bacterial pathogens in blood. Comp Immunol Microbiol Infect Dis 32, 207-219. [2] Disqué C (2007) Einfluss der DNA-Extraktion auf die PCR-Detektion von Sepsiserregern. BIOspektrum 06, 627-629. (in German) [3] Krohn S, Böhm S, Engelmann C, Hartmann J, Brodzinski A, Chatzinotas A, Zeller K, Prywerek D, Fetzer I, Berg T (2014) Application of qualitative and quantitative real-time PCR, direct sequencing, and terminal restriction fragment length polymorphism analysis for detection and identification of polymicrobial 16S rRNA genes in ascites. J Clin Microbiol 52, 1754-1757. [4] Rudkjøbing VB, Aanaes K, Wolff TY, von Buchwald C, Johansen HK, Thomsen TR (2014) An exploratory study of microbial diversity in sinus infections of cystic fibrosis patients by molecular methods. J Cys Fibrosis doi:10.1016/j.jcf.2014.02.008. [5] Thoendel M, Jeraldo PR, Greenwood-Quaintance KE, Yao JZ, Chia N, Hanssen AD, Abdel MP, Patel R (2016) Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing. J Microbiol Meth 21, 141-145. [6] Xu Y, Larsen LH, Lorenzen J, Hall-Stoodley L, Kikhney J, Moter A, Thomsen TR (2017) Microbiological diagnosis of device-related biofilm infections. APMIS 125, 289-303. |
| Application notes |
| MolYsis-SelectNA™plus Micro-Dx™ |

 Molecular analysis of microbial DNA in primary sterile clinical samples like whole blood is challenging. Human DNA is found in up to several thousand-fold mass excess to bacterial or fungal DNA and tends to bind primers unspecifically [1]. Consequently, unspecific primer binding entails loss in analytical sensitivity and specificity, particularly when the microbial load is low [2].
Molecular analysis of microbial DNA in primary sterile clinical samples like whole blood is challenging. Human DNA is found in up to several thousand-fold mass excess to bacterial or fungal DNA and tends to bind primers unspecifically [1]. Consequently, unspecific primer binding entails loss in analytical sensitivity and specificity, particularly when the microbial load is low [2].