MolYsis™ Host DNA Removal – Effect on Pathogen Analysis |
|
| 09 April 2018 | |
 PCR and real-time PCR are used as sensitive methods for the qualitative and quantitative detection of microorganisms in medical and pharmaceutical applications, including QC in production management, infection diagnosis, as well as in patient surveillance and monitoring. Cell cultures and clinical specimens contain large amounts of host DNA that often exceed microbial DNA several thousand fold. In low microbial load samples, primers specific for target sequences like the broadly used 16S or 18S rRNA genes of bacteria and fungi, respectively, tend to bind to host DNA unspecifically. PCR and real-time PCR are used as sensitive methods for the qualitative and quantitative detection of microorganisms in medical and pharmaceutical applications, including QC in production management, infection diagnosis, as well as in patient surveillance and monitoring. Cell cultures and clinical specimens contain large amounts of host DNA that often exceed microbial DNA several thousand fold. In low microbial load samples, primers specific for target sequences like the broadly used 16S or 18S rRNA genes of bacteria and fungi, respectively, tend to bind to host DNA unspecifically. As a result, the DNA polymerase is mainly engaged in the synthesis of single and occasionally double-stranded host sequences. In quantitative real-time PCR assays, this is seen as a shift of the threshold to higher C(t) values and lower limit of detection compared to pure bacterial DNA [1]. In gel electrophoresis, amplicons from host DNA appear as multiple bands in addition to bacteria specific signals. |
|
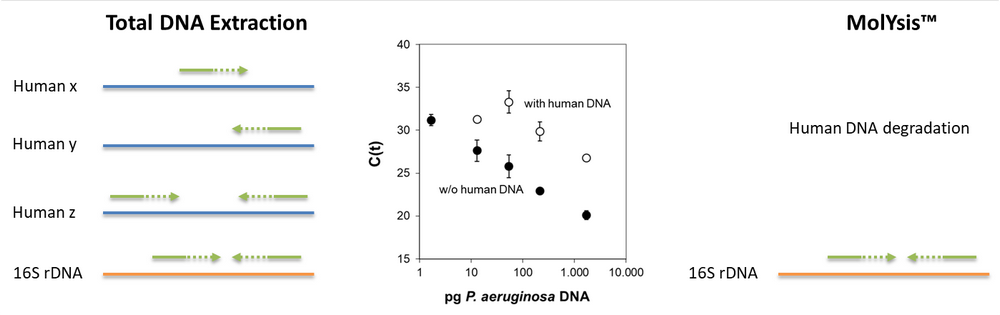 |
|
| Fig.1 Unspecific binding of primers to human DNA (left sketch) limits their availability for specific amplification of target microbial sequences. MolYsis™ degrades the human DNA before extraction of microbial DNA (right). In quantitative real-time PCR, threshold values are lower without than with human DNA (central graph; after [3]). Degradation of human DNA thus increases the sensitivity of the assay. | |
| Human DNA degradation enhances amplification-based assay accuracy | |
| Loss of sensitivity and false positive results in PCR-based assaying are the effects of primer – host DNA interaction. Molzym’s MolYsis™ technology involves selective lysis of host cells while microorganisms, because of their rigid cell walls, stay intact. Subsequently, the released host DNA is degraded by a DNase and, at the end, the microorganisms are lysed and the DNA is purified. The clear advantage of this technique is demonstrated in a recent NGS comparative study [2]. Metagenomic whole genome sequencing of resected arthroplasty component sonicate fluid revealed 481 to 9580-fold enrichment of reads from the pathogens known to be in the samples compared to unenriched DNA extracts. Apparently, the removal of the vast amount of host sequences led to a considerable increase in target reads in this next generation sequencing approach. MolYsis™ has been used in a variety of other applications which are discussed in a recent application note [3]. | |
| References | |
| [1] Handschur M, Karlic H, Hertl C, Pfeilstöcker M, Haslberger AG (2009) Preanalytic removal of human DNA eliminates false signals in general 16S rDNA PCR monitoring of bacterial pathogens in blood. Comp Immunol Microbiol Infect Dis 32, 207-219. (link) | |
| [2] Thoendel M, Jeraldo PR, Greenwood-Quaintance KE, Yao JZ, Chia N, Hanssen AD, Abdel MP, Patel R (2016) Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing. J Microbiol Meth 21, 141-145. (link) | |
| [3] Linow M (2018) Applications of MolYsis™ host DNA removal in culture-independent molecular analysis of bacteria and fungi. Res Mol Microbiol 1/18, 1-3. | |
| Brochure Application Note | |
