Diagnosis of Rare Pathogens by Direct 16S Sequencing |
|
| 23 May 2019 | |
|
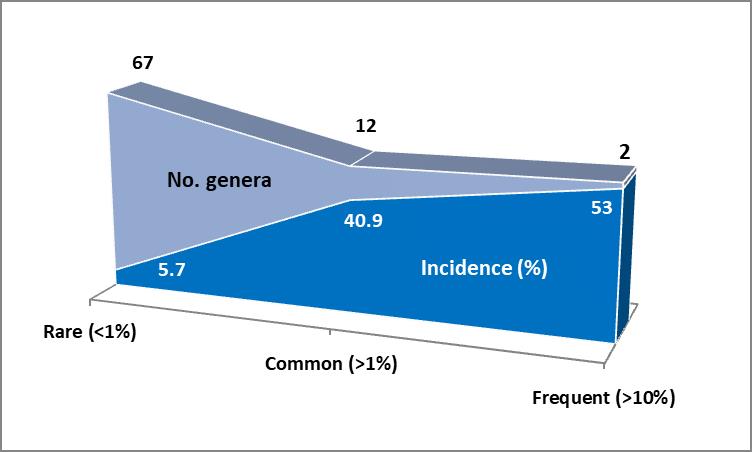
In his whitepaper, Lorenz [1] addresses the diversity of bacterial pathogens of the systemic system leading to sepsis and septic shock. A survey of five large clinical studies performed in Europe, Asia and North America shows that there are only three bacteria from two genera, Staphylococcus aureus, other staphylococci and Escherichia coli, that in the majority (median: 53%) are the cause of infection (Fig. 1). Diversity increases markedly (12 genera) with other, less frequent (40.9%) but common pathogens. When taken together results from the five large studies and a survey of case studies, the most diverse group (67 genera) comprises of bacteria that are occasionally found (< 1% of cases). This list is not claimed to be comprehensive, but the trend clearly points out that conventional microbiological diagnosis may face limitations by unusual growth conditions and phenotypic markers for the differentiation and identification of rare bacteria that stem from a variety of habitats apart from the human body and belong to very different phylogenetic and physiological groups.
Fig. 1: Diversity of bacterial pathogens involved in sepsis. Incidence: median of the summed up incidences of pathogens found in studies from 5 countries in the groups ‘frequent’ (>10%), ‘common’ (≥1%) and ‘rare’ (<1%). Number of genera of rare and unusual bacteria found in the studies above and case studies. See [1] for the full list of pathogens. Lorenz [1] emphasises the benefits of rRNA gene sequencing as an accepted and precise means of strain identification. Especially for the diverse group of rare pathogen which are potentially unidentifiable by conventional microbial diagnosis, rRNA gene sequencing creates an added value. A consequence of this would be to use 16S rRNA gene sequencing in an attempt to identify a broad range of bacteria in a direct, i.e., culture-independent way. There is plenty evidence that Molzym’s manual and automated CE IVD-marked products, SepsiTest™-UMD and Micro-Dx™, provide standardised tools to fulfil the requirements of a sensitive and specific sequencing-based method for the direct in-vitro diagnosis of hundreds of pathogens in a variety of clinical materials – using only a single protocol. |
|
| Reference | |
| [1] Lorenz MG (2019) Rare pathogens – culture-independent eubacterial 16S rRNA gene diagnosis. Trends in Molecular Diagnostics, no. 1/19: 1-9 (link). | |