Remove Host DNA and Use Your Validated Extraction System! |
| 10 September 2018 |
|
DNA extraction of clinical samples from primary sterile body sites and other specimens by the methods established often results in eluates that contain human DNA in vast amounts and only sparse amounts of target microbial DNA. Under this condition, human DNA tends to bind primers designed for the PCR amplification of microbial sequences. The effect of such unspecific primer binding is a loss of sensitivity and specificity (blog post). Further, the vast majority of reads in NGS whole genome sequencing comes from human sequences and leads to low resolution of microbiome reads, respectively. With MolYsis™ basic, Molzym offers a solution to the problem without the necessity of changing the established DNA extraction system. |
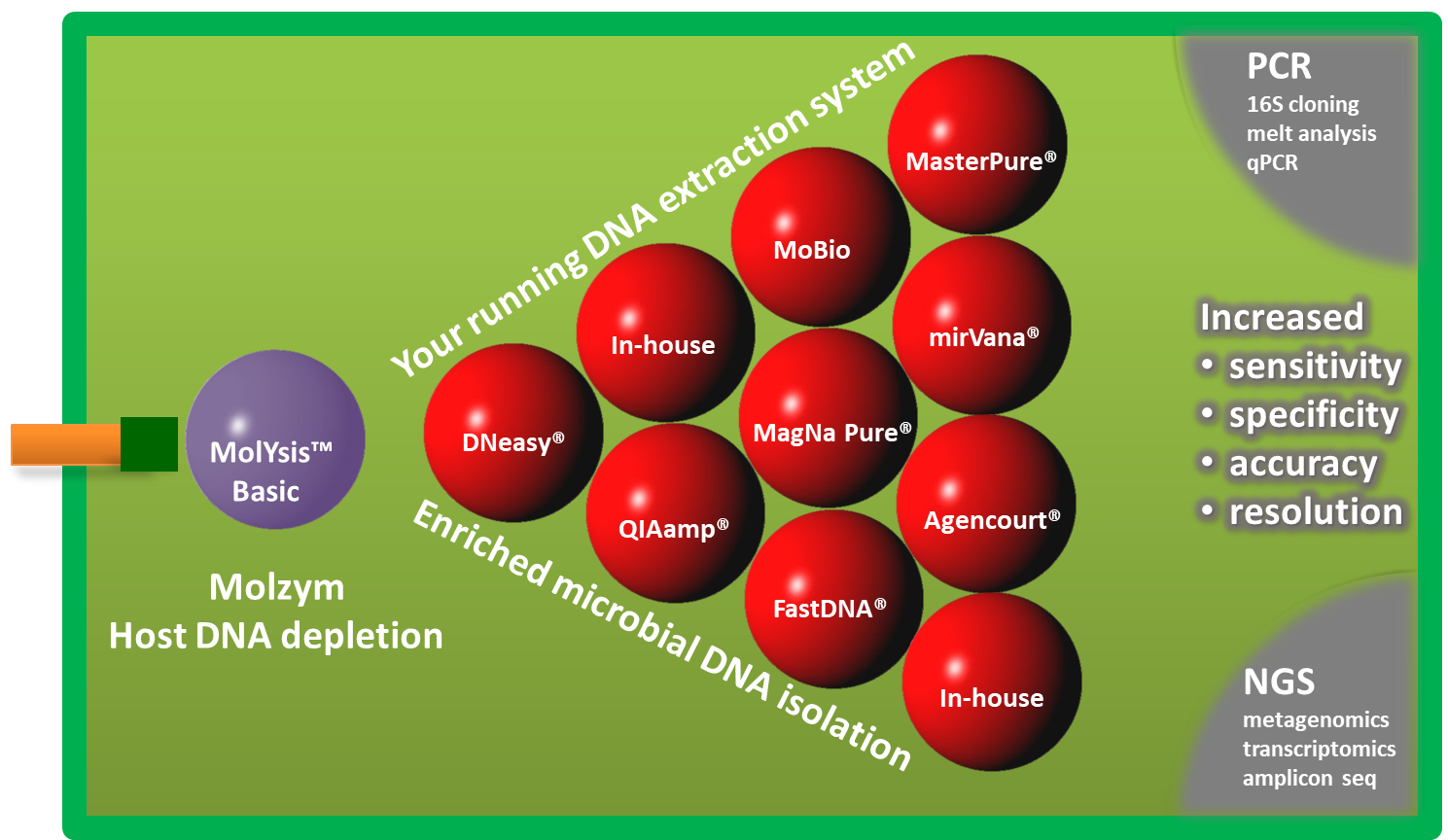 |
|
MolYsis™ Basic kits enable the depletion of human DNA up to 99% from samples of 0.2 to 10ml volume. At the end of the short treatment, enriched microbial cells are steered into any manual or automated DNA extraction system that is being used routinely in your laboratory. MolYsis™ Basic has proven its superior performance with a series of applications, including PCR, Real-Time PCR, qPCR, high resolution melt analysis, cloning, metagenomics, transcriptomics and amplicon resequencing approaches [1-11]. Call for more information at: +49 421 69616215 |
| References |
|
[1]] Xu Y, Børsholt Rudkjøbing V, Simonsen O, Pedersen C, Lorenzen J, Schønheyder HC, Nielsen PH, Rolighed Thomsen T (2012) Bacterial diversity in suspected prosthetic joint infections: an exploratory study using 16S rRNA gene analysis. FEMS Immunol Med Microbiol 65, 291–304. [2] Fricke WF, Maddox C, Song Y, Bromberg JS (2014), Human microbiota characterization in the course of renal transplantation. Am J Transplant 14, 416–427. [3] Thoendel MJ, Jeraldo PR, Greenwood-Quaintance KE, Yao JZ, Chia N, Hanssen AD, Abdel MP, Patel R (2018) Identification of prosthetic joint infection pathogens using a shotgun metagenomics approach. Clin Inf Dis doi.org/10.1093/cid/ciy303. [4] Masek BJ, Hardick J, Won H, Yang S, Hsieh YH, Rothman RE, Gaydos CA (2014) Sensitive detection and serovar differentiation of typhoidal and nontyphoidal Salmonella enterica species using 16S rRNA Gene PCR coupled with high-resolution melt analysis. J Mol Diagn 16, 261–266. [5] Dekker JP (2018) Metagenomics for clinical infectious disease diagnostics steps closer to reality. J Clin Microbiol doi:10.1128/JCM.00850-18. [6] Rudkjøbing VB, Aanaes K, Wolff TY, von Buchwald C, Johansen HK, Thomsen TR (2014) An exploratory study of microbial diversity in sinus infections of cystic fibrosis patients by molecular methods. J Cys Fibrosis 13:645-652. [7] Zhao Y, Armeanu E, DiVerniero T, Lewis TA, Dobson RC, Kontoyiannis DP, Roilides E, Walsh TJ, Perlin DS (2014) Fungal DNA detected in blood samples of patients who received contaminated methylprednisolone injections reveals increased complexity of causative agents. J Clin Microbiol 52, 2212-2215. [8] Duran-Pinedo AE, Chen T, Teles R, R Starr JR, Wang X, Krishnan K, Frias-Lopez J (2014) Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. The ISME J 8:1659-1672. [9] Ivy MI, Thoendel MJ, Jeraldo PR, Greenwood-Quaintance KE, Hanssen AD, Abdel MP, Chia N, Yao JZ, Tande AJ, Mandrekar JN, Patel R (2018) Direct detection and identification of prosthetic joint infection pathogens in synovial fluid by metagenomic shotgun sequencing. J Clin Microbiol. doi: 10.1128/JCM.00402-18. [10] Peterson SW, Knox NC, Golding GR, Tyler SD, Tyler AD, Mabon P, Embree JE, Fleming F, Fanella S, Van Domselaar G, Mulvey MR, Graham MR (2016) A study of the infant nasal microbiome development over the first year of life and in relation to their primary adult caregivers using cpn60 universal target (UT) as a phylogenetic marker. PLoS ONE 11, e0152493. doi:10.1371/journal.pone.0152493. [11] Linow M (2018) Applications of MolYsis™ host DNA removal in culture-independent molecular analysis of bacteria and fungi. Res Mol Microbiol 1/18, 1-3 (download). |
| Brochure Product Information |

 Standardisation is important for the generation of reliable results in research and routine molecular infection diagnosis. DNA extraction is an important pre-analytical step on the way to pathogen analysis. The establishment of certain in-house methods or commercial kits in the laboratory is the result of long-term validation processes.
Standardisation is important for the generation of reliable results in research and routine molecular infection diagnosis. DNA extraction is an important pre-analytical step on the way to pathogen analysis. The establishment of certain in-house methods or commercial kits in the laboratory is the result of long-term validation processes.